Getting Started
1. Intake Form
The PPMS intake form is required for all new data requests. If you are a collaborator of Dr. Madadbhushi or Brain Health, please create a placeholder dummy account number.
2. Data Requirements for Successful Extraction
Please review this section carefully. For the effective and accurate extraction of DICOM images, specific data elements are required. Failure to provide required data will result in delays in extraction.
a. Radiology Accession Numbers (Preferred): These are the preferred identifiers for extracting images, as they are specific to each exam and directly linked to the images that need to be extracted. Using accession numbers minimizes the risk of retrieving unnecessary or incorrect data. If accession numbers are provided, no other information is required to retrieve exams. If you do not have access to accession numbers, they can be requested through the data solutions team using this form without any additional cost.
Examples:
E01234567
0001CT2212345678 (16-digit)
0001MR202212345678 (18-digit)
b. Epic ID/EMPI + Specific Date: When retrieving specific studies, a combination of Epic ID (previously EMPI) and a specific date may be provided to extract the requested studies.
An important note on the relationship between Epic IDs, EMPI, and MRNs:
The Epic ID (previously EMPI) is a unique identifier for patients across all Emory hospitals and is the identifier stored in radiology PACS. Therefore, it is required for image extraction unless accession numbers are available. Historically, the EMPI was developed to unify patient identification across the enterprise after the integration of multiple hospitals (ECLH, SJH, EJCH, DOU) that initially had their own medical record numbers (MRNs) which could often overlap, leading to ambiguity. These hospital-specific MRNs (ECLH-MRN, TEC-MRN, EJCH-MRN, EUH-MRN, SJH-MRN, DOU-MRN) cannot be used for extraction. Conversion of MRN to EMPIs can be requested through the data solutions team using this form without any additional cost.
c. Epic ID/EMPI + Date Range: For relatively small numbers of patients and narrow date ranges, a combination of Epic ID/EMPI and range of dates can be used to retrieve studies. Generally, if the query must be < 500 patients and a date range of <90 days to be feasible. Each patient ID and date combination is a unique query, i.e. querying 500 patients in a 90-day range results in 45,000 queries to PACS. Any studies that fall within the date range will be retrieved, and this tends to yield datasets that need to be cleaned after extraction. Modality can be added for further filtering (see #d).
d. Modality: When specific accession numbers are not available, the modality can help further refine queries based on patient ID and date. You may specify the type(s) of scans requested, for example CT, MR, US, XR, NM, or PT. Specific anatomies (for example, CT head or XR Chest) cannot be requested directly and must be filtered out after all exams of the modality are retrieved.
3. Output Directory Structure
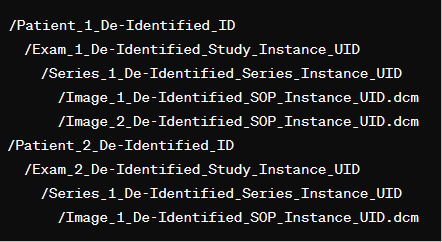
The output format for the extracted data is organized into a hierarchical directory structure that mirrors the structure of PACS: /PatientID/ StudyUID/ SeriesUID/ SOPInstanceUID.dcm.
Following de-identification, original patient IDs and Study/Series/Instance UIDs are replaced with their de-identified values. Each patient folder contains subfolders for their exams, which in turn contains subfolders for each series of images from that exam, and finally, the individual DICOM images are stored within these series folders.
Below is an example of how the data is structured.
4. Output Metadata File
Alongside the image data, a metadata CSV file (metadata_anon.csv) is provided, which contains select de-identified DICOM metadata for each image. This includes de-identified patient IDs, accession numbers, dates, as well as original (unmodified) technical parameters. The metadata file allows easily cataloging of files contained in the extract and enables identification of images that may meet a certain criteria – for example, an exam type or manufacturer. The de-identified DICOM files will contain more complete metadata and can be opened and parsed using packages like pydicom as needed.
Key Columns in Metadata_anon.csv:
- PatientID_anon
- AccessionNumber_anon
- StudyDate_anon
- StudyInstanceUID
- SeriesInstanceUID
- SOPInstanceUID
- Modality
- Manufacturer
- ManufacturerModel
- Dose
- FieldStrength
- StudyDescription
- SeriesDescription
- ImageLaterality
- ViewPosition
Linking Table
To link de-identified data (patient ID, accession numbers, and dates) with original identifiers (for authorized purposes only), a linkage file can be provided. This file contains mappings between original and anonymized identifiers, which can be used for audit trails or to link patients across datasets:
Contents of Linkage File:
- PatientID ↔ PatientID_anon
- AccessionNumber ↔ AccessionNumber_anon
- PatientID ↔ date shift (+/- days)
The linkage file is carefully controlled and only made available under strict conditions to preserve the integrity of the de-identification process.
These output formats and supplementary files ensure that while the data remains fully usable for research and analysis purposes, it adheres strictly to privacy standards and regulations.