Resources
General
How to use FlowJo off-campus using an Emory VPN connection
Follow these instructions to create and open a VPN (Virtual Private Network) connection to the Emory system.
Then fire up FlowJo (make sure you've registered FlowJo!). Let us know if you have any questions.
Protocols and Procedures
- Antibody Titering ( Carleton Stewart, et al)
- Immunophenotyping (Carleton Stewart, et al)
- Cell Cycle Staining Protocol (special Thanks to Dr. Ritu Aneja and the Harish Joshi Lab)
- A WinMDI Tutorial
- A Setup System for Compensation: BD CompBeads plus BD FACSDiva Software (pdf)
- Determining Antibody Titers (special Thanks to Dr. Carleton Stewart and RPCI)
- Titering Directly Conjugated Antibodies to Extracellular Antigens (special Thanks to Dr. Carleton Stewart and RPCI)
- Apoptosis Prep for Training
- Extracellular Immunophenotyping prep (special Thanks to Hilary Rosenthal, MT (ASCP) and the Waller Lab)
- Intracellular Staining prep (special Thanks to Hilary Rosenthal, MT (ASCP) and the Waller Lab)
- Cell-cycle protocol for yeast (special Thanks to Missy Brykailo and the Fridovich-Keil Lab)
Training Materials
- BD FACSymphony™ A3 Flow Cytometer User’s Guide
- BD FACSymphony™ A5 SE Flow Cytometer User's Guide
- Cytek Aurora User’s Guide
Sorting
In order to avoid the common pitfalls inevitable in preparing cells for sorting, we recommend reading the following links:
Sorting Presentations
- How a sorter works:
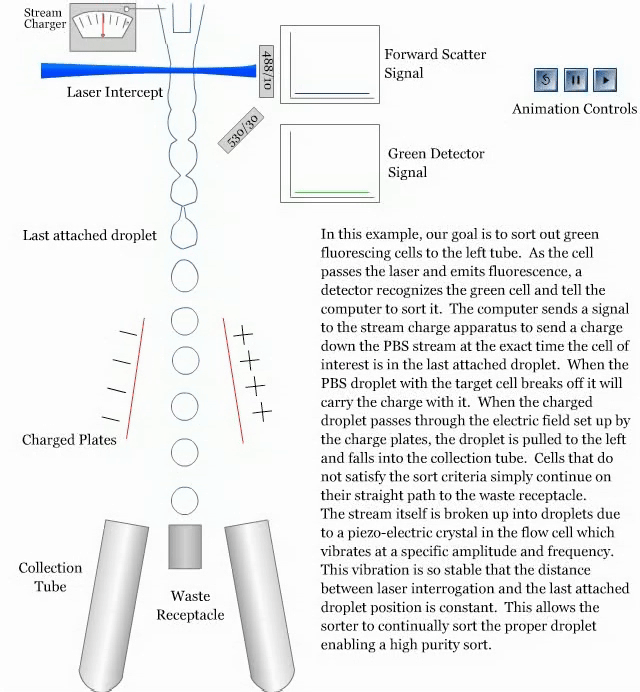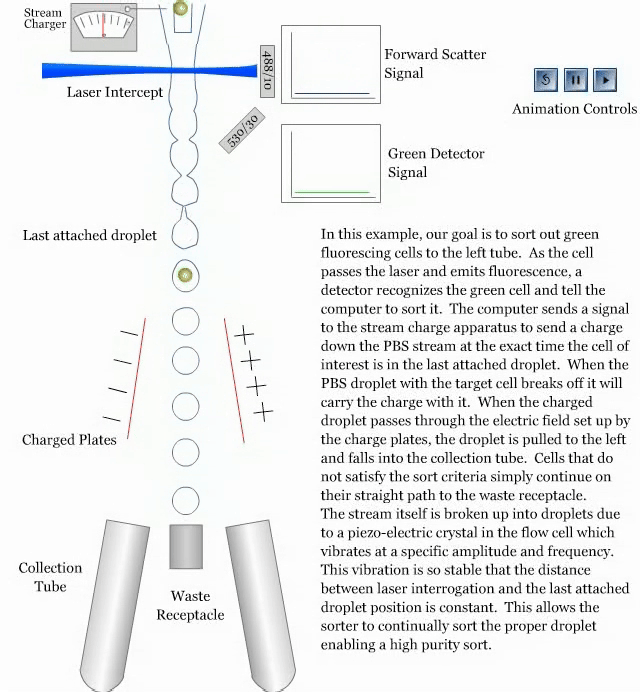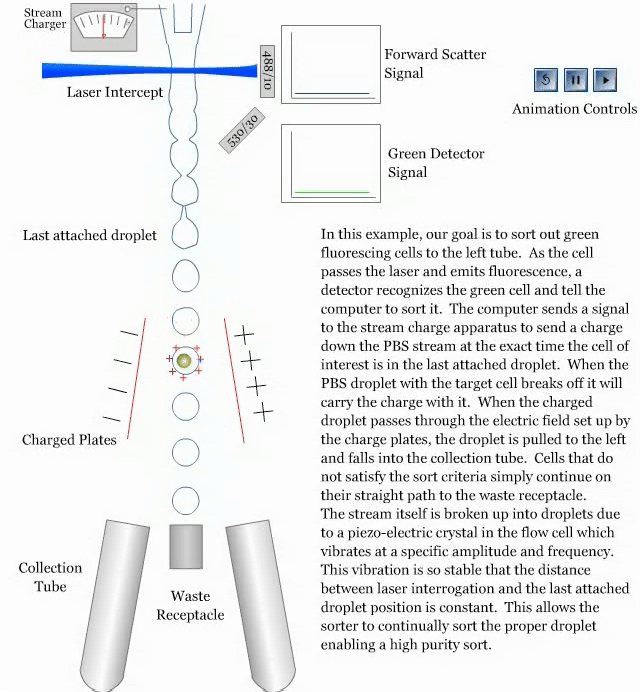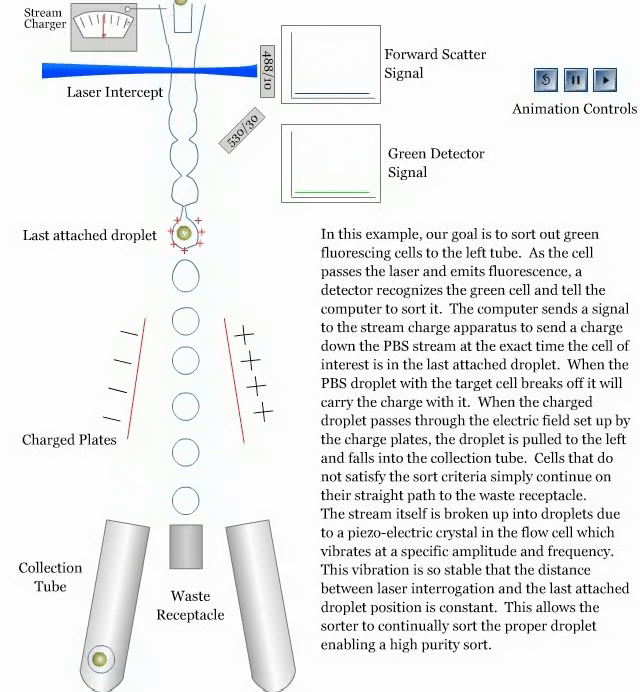
References
- Flow Cytometry: A Practical Approach. Edited by MG Ormerod. (IRL Press, Oxford. 1994. ISBN 0-19 963461-0)
- Practical Flow Cytometry. 3rd Edition. Howard M Shapiro. (Alan R Liss, Inc. ISBN 0-471-30376-3).
- Flow Cytometry. First Principles. Alice Longobardi Givan. (Wiley-Liss, New York, 1992. ISBN 0-471-56095-2.)
- Handbook of Flow Cytometry Methods. Edited by J Paul Robinson. (Wiley-Liss, New York, 1993. ISBN 0-471-59634-5.)
Facility Description
The Emory School of Medicine Core Facility for Flow Cytometry (EFCC) provides analysis and sorting services and experimental/panel design to the research community in & around Emory University Campus (Emory, Georgia Tech, & UGA). Established in 2001, the SOM Flow Core offers a wealth of experience & knowledge relative to Flow Cytometry.
Centrally Located within the heart of Emory’s Health Science & Research Quad (O. Wayne Rollins Research Center, Whitehead Biomedical Research, Emory Vaccine Center, Rollins School of Public Health & HSRBII), we offer a convenient location staffed with dedicated professionals with decades of experience.
Instrumentation
Analysis:
- Symphony A3, A5 SE* (Becton Dickinson),
- Aurora* (Cytek),
- 24+ Fluorescence Detectors
- Lasers
- UV (355 nm)
- Violet (405 nm)
- Blue (488 nm)
- Green (561 nm)
- Red (640 nm)
Sorters:
- FACSymphony S6 SE*, FACSDiscover S8* (Becton Dickinson)
- Aurora CS* (Cytek)
- 24+ Fluorescence Detectors
- Lasers
- UV (355 nm)
- Violet (405 nm)
- Blue (488 nm)
- Green (561 nm)
- Red (640 nm)
Sort Capabilities:
- 6-way Sorting
- Single Cell Plate & Index Sorting
- Temperature Controlled Sorting Into:
- Tubes
- 6, 24, 48, 96, & 384 well plates
- Sorting onto Slides
- Imaging (FACSDiscover S8)