µCourse - BioImage Analysis @ Emory
A quantitative data analysis course that included a weekly mixture of lectures and workshops. Course information and recordings are here for you to explore.
Join our Slack channel bia-at-emory at emoryici.slack.com, to ask questions
- How do I ask questions or contact with any questions during the course?
We will use our slack channel bia-at-emory on emoryici.slack.com workspace for answering questions. Most likely, if you have a question someone else has it too.
- I am having issues accessing the slack channel
Email us at ici@emory.edu and we will send you a personal invite you to access the channel. Only institutional email addresses will be granted access.
- I do not have any experience with Image analysis. Is this course a right fit for me?The course requires no prior experience with image analysis.
- I do not perform microscopy experiments. Is this course a good time investment?
This course will help you think or plan for imaging based experiments with an end goal of image quantification.
- Are Image J and Fiji the same thing?
Fiji is just Image J (Fiji), but with all the essential plugins integrated to the software (ImageJ2), and an ease of installing plugins from different sites. The community prefers writing "Fiji" over "FIJI". More technical information on differences in the architecture of ImageJ can be found at https://imagej.net/
- Where do I download Fiji from?
https://fiji.sc/
- Where do I download CellProfiler from?
https://cellprofiler.org/releases
- CellProfiler does not run on my PC?
Follow this thread and threads within on Image.sc forum
- Can I join for few workshops and skip others?
The workshops are linked to one another. It is highly recommended to attend them sequentially.
- Do I need to have any coding experience?
No. This course will use softwares (Fiji, CellProfiler, and other deep learning tools) with graphical user interface.
- Does this course require registration?
This is not a formal course that requires registration.
- Is there a fee for this course?
There is no fee for this course.
- Still any questions?
Email us at ici@emory.edu
This course will help you
- Design experiments with quantification as an end goal
- Use Fiji to perform basic image operations on image acquired from any of the core microscopes and beyond
- Segment images using Fiji and deep learning models
- Use macro language in Fiji to automate repetitive tasks
- Segment objects either using pre-existing models or training models with deep learning approaches
- Edit and create Cell Profiler pipeline to analyze data
- Integrate Fiji, deep learning tools (such as Cellpose and StarDist), and Cell Profiler to analyze and quantify hard to segment datasets
- Get an overview on some of the tools available in the constantly evolving field of Deep Learning
- Analysis and application of high content screening data and its role in drug discovery process
- Create awareness of 3D and 4D data visualization tools such as Imaris and Vison 4D
- Understand and navigate Colocalization
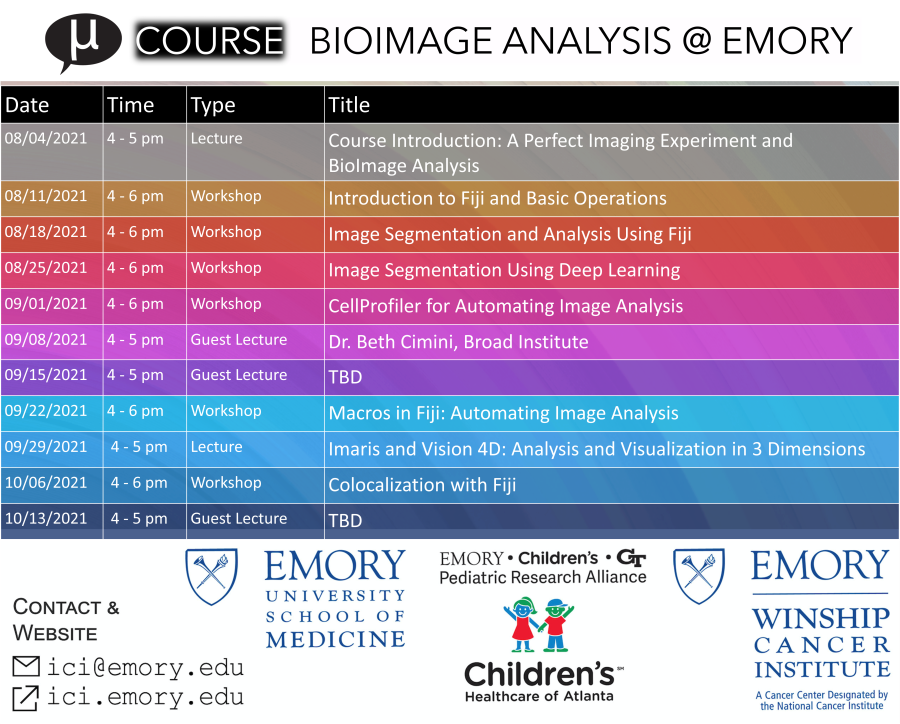
SCHEDULE
Course Introduction: A Perfect Imaging Experiment and BioImage Analysis | ||
Introduction to Fiji: Basic Operations | ||
Image Segmentation and Analysis using Fiji | ||
Image Segmentation Using Deep Learning | ||
CellProfiler for Automating Image Analysis | ||
Making the most of your microscopy data with high content analysisDr. Beth Cimini, Broad Institute | ||
Deep Learning for microscopy image analysis Dr. Sreenivas Bhattiprolu, Zeiss Microscopy | ||
Automation with Fiji: Intro to Macros | ||
Imaris and Vision 4D: Analysis and Visualization IN 3 DIMENSIONS | ||
Colocalization with Fiji |